Research in the Morris Lab




An overview of our research
Cell fate is flexible
The generation of clinically relevant cells, such as neurons, cardiomyocytes, and hepatocytes, in vitro offers potential for regenerative therapy and permits disease modeling, toxicology testing, and drug discovery. Cell differentiation had long been thought a unidirectional process toward restricted potential and increased specialization. In the past half-century, this has been challenged: Mature somatic cells can be returned to a pluripotent state, and subsequently differentiated to desired cell types. Alternatively, mature cells can be ‘directly converted’ from one mature state to another via transcription factor overexpression, bypassing pluripotency. Many approaches are employed to generate defined fate in vitro, however, the resultant cells often appear developmentally immature or incompletely specified, limiting their utility.
The challenges of assessing and engineering cell identity
The evaluation of cell fate has been confounded by the lack of any systematic means by which to assess the fidelity of engineered cells. We were involved in the development of ‘CellNet’, a network biology-based computational platform that accurately evaluates cell fate through gene regulatory network reconstruction and generates hypotheses for improving cell differentiation protocols (see Cahan et al., Cell, 2014, and Morris et al., Cell, 2014). Using this platform we surveyed a range of engineered cells and found that cells derived via directed differentiation more faithfully recapitulated target cell identity than cells generated by direct conversion. These directly converted cells commonly failed to silence expression programs of the original cell type, and illicit gene expression programs were frequently induced. Employing induced endoderm progenitors (iEPs) generated from fibroblasts as a prototypical conversion, our computational and functional analyses showed that iEPs behave as embryonic progenitors with the potential to functionally engraft both the liver and colon. We found that these engineered cells resembled mature colonic epithelium after transplantation into the colon niche.
Now, we are investigating how the transcription factors utilized in these conversion protocols impact the GRN of the host cell to drive changes in cell identity. We are particularly interested in pioneer factors, capable of remodeling chromatin and often re-purposed during development. To study the mechanism of conversion we use a combination of transcriptional, epigenetic, and functional analyses. This understanding will help us guide the activity of conversion factors and unlock further progenitor potential of engineered cells. In addition, this line of investigation will be broadly applicable to the cell fate engineering field and may also shed light on the activity of transcription factors during development.
Single-cell mapping of lineage and identity in cell fate reprogramming
Recently, we have revealed further mechanisms of reprogramming to iEPs. Successful reprogramming is a rare event, thus it had remained a challenge to isolate and analyze the few iEPs emerging from fibroblasts. High-throughput single-cell RNA-sequencing has enabled this heterogeneity to be deconstructed, although lineage relationships between cells were lost, making interpretation of the data a challenge. To overcome this, we have developed a straightforward, high-throughput cell tracking method, ‘CellTagging’ (see Biddy et al., Nature, 2018). Sequential lentiviral delivery of heritable random unique molecular indexes, CellTags, permits the construction of multi-level lineage trees. CellTagging and longitudinal tracking of fibroblast to induced endoderm progenitor reprogramming revealed two distinct trajectories: one leading to successfully reprogrammed cells, and one leading to a ‘dead-end’ state, paths determined in the earliest stages of lineage conversion. We found that expression of a putative methyltransferase, Mettl7a1, is associated with the successful reprogramming trajectory; adding Mettl7a1 to the reprogramming cocktail increased the yield of induced endoderm progenitors.
We continue to focus on the development of new single-cell technologies to track cell identity (see Guo et al., BioRxiv, 2018). Our main focus is to apply these tools to a broad range of reprogramming systems to uncover the general mechanisms of cell fate conversion. In addition, we are tracking the maturation of iEPs in the intestine at the single-cell level following transplantation. Mapping these in vivo differentiation steps will help guide maturation in vitro and also assist in identifying factors to promote liver repopulation and maturation. This line of investigation will aid in answering important questions about silencing of the donor cell program and establishment of the target cell GRNs.
Images from the lab
CellTagging lineages
Reconstruction and visualization of lineages undergoing fibroblast to induced endoderm progenitor reprogramming. Each node represents an individual cell, and edges represent clonal relationships between cells. Clonal relationships were reconstructed from each cell’s CellTag expression profile. The different colors denote times of sequential CellTag labeling.
Cell Cover T Map
The cover depicts an adaptation of the Boston subway system map in which subway lines represent pathways of differentiation from pluripotent stem cells. Bus routes, shown as dashed lines, are the various ways that cells can be experimentally interconverted via directed differentiation, direct conversion, and reprogramming.
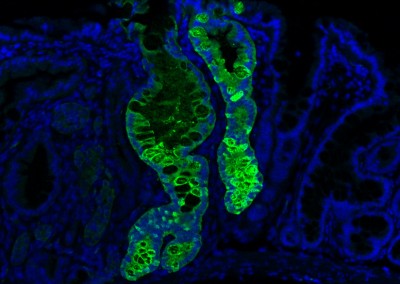
Engrafted Colon
Long-term engrafted GFP-iHeps/iEPs in a section of colon seven weeks after transplantation. GFP-labelled iHeps integrate into the damaged colonic epithelium to functionally engraft the tissue.
Engrafted Colon
Alkaline Phosphatase staining of long-term engrafted GFP-iHeps/iEPs (brown) in a section of colon seven weeks after transplantation. Goblet cells are visualized by PAS staining (purple). GFP-labelled iHeps integrate into the damaged colonic epithelium to functionally engraft the tissue.
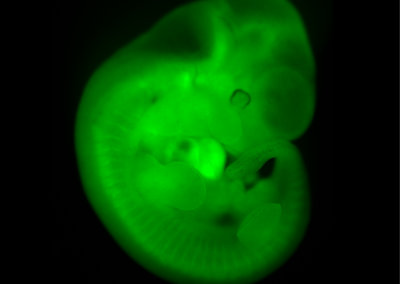
Chimeric Mouse Embryo
E11.5 chimeric embryo derived from a wild-type CBAxC57/BL6 embryo and Histone2B-Green Fluorescent Protein transgenic embryonic stem cells. The ES cells contribute to all tissues of the embryo.
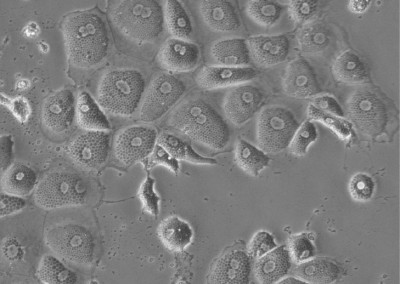
Induced Hepatocytes
Bright-field image of induced hepatocytes (iHeps) in culture. Fibroblasts were directly converted to iHeps through forced expression of the transcription factors, Foxa1 and Hnf4a.
Mouse ESCs
Mouse Embryonic Stem Cells derived from Histone2B-Green Fluorescent Protein transgenic embryos.
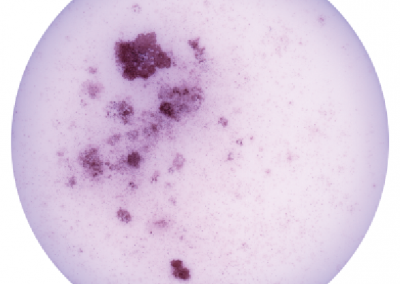
Induced Hepatocyte Colonies
Alkaline Phosphatase staining of E-cadherin expressing Induced Hepatocyte colonies, three weeks following their direct conversion from fibroblasts, driven by the transcription factors Foxa1 and Hnf4a.
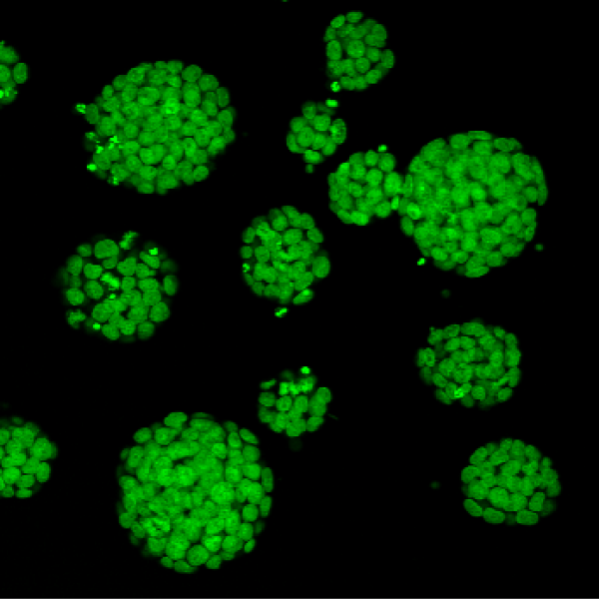
Colonosphere
Bright-field image of Induced hepatocytes (iHeps) cultured as three dimensional organoids in matrigel.
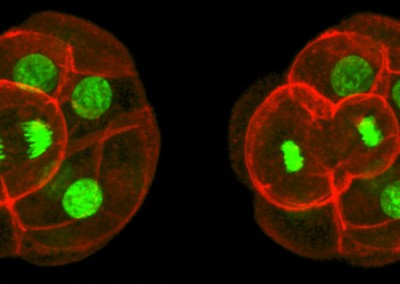
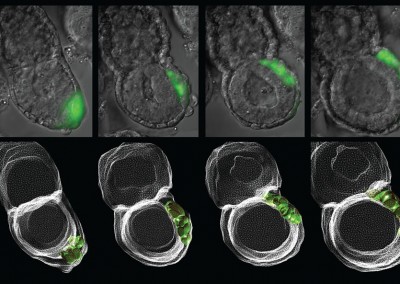
Asymmetric Cell Division
Confocal imaging time-lapse series of an asymmetrical cell division in an embryo transitioning from the 8-cell to 16-cell stage during early development of an Histone2B-Green Fluorescent Protein transgenic embryo expressing a membrane-targeted form of Red Fluorescent Protein.
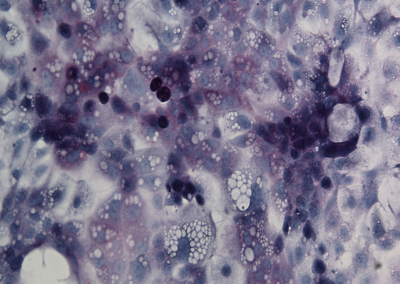
iHep Glycogen Storage
Glycogen is made and stored primarily in the cells of the liver and the muscles, and functions as the secondary long-term energy storage. Here, glycogen storage in iHeps directly converted from fibroblasts via expression of the transcription factors Foxa1 and Hnf4a is visualized by a Periodic acid-Schiff stain (purple).
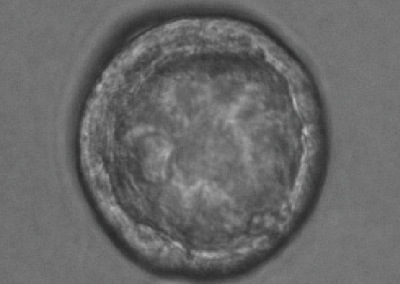
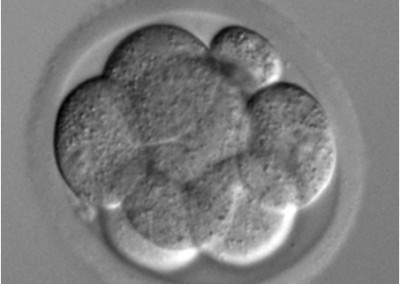
8-cell stage embryo
Phase contrast microscope image of an 8-cell stage pre-implantation mouse embryo cultured in vitro.
Mouse Blastocyst
Confocal fluorescent image of a mouse embryo in the late stages of pre-implantation development, just prior to implantation into the uterus. Cell membranes are visualized in green and the Primitive Endoderm is visualized by staining Sox17a in red. At these late stages, the blastocyst has begun hatching from the Zona Pellucida.